Clubfoot is a complex disorder meaning that more than one gene as well as environmental factors will be discovered to play a role in its etiology. Identifying the genes for clubfoot will allow for improved genetic counseling and may potentially lead to new and improved treatment and preventive strategies for this disorder.
FAMILY
of all patients with isolated clubfoot report a positive family history for this defect
ENIGMA
You surely ask yourself the same question as every parent of a child born with a congenital clubfoot? You’re looking back on your pregnancy memories, trying to find a clue which could give you an answer for the question that still remains a mystery… The research on the cause of clubfoot vary has continued ceaselessly for many years, bothering many researchers around the world. Geneticists don’t stop looking for this ‘magical’ gene which is the cause of this foot defect. Unfortunately, it is known nowadays that there is more than one different cause, which makes the investigation much more complicated, the research has not yet determined the root cause. Clinicians have made countless hypotheses concerning the involvement of genetic factors, however many of them contradict each other. Thus, in the history of the research we can find a theory of: an autosomal recessive inheritance (Fetscher (1921), Idelberger (1939), J. A. Böök (1948), Ducci and Grilli (1954)), an autosomal dominant inheritance with reduced penetrance (R. Palmer (1964)) or a sex-linked recessive inheritance (E. Isigkeit (1927)).
Constantly improving researching methodes and science development lead the researchers to the conclusion that as far as single genes which could possibly participate in inheritance are important, a polygenic inheritance cannot be excluded. It was mentioned by many scientists, for example Wynne-Davies in 1965 or above mentioned Palmer in 1974 or Yamamoto in 1979. Today we know that clubfoot defect a is a heterogeneous disorder (including many factors) with a polygenetic threshold, meaning that many genes participate in its formation. The discovery of these participating genes and the understanding of formations of certain gene anomalies and their interaction with other genes are the goal of current research.
THE DISCOVERY FROM ST. LOUIS – PITX1 AND TBX4
In 2008, a group of scientists (including Dr Matthew B. Dobbs) from the Washington University School of Medicine in St. Louis, USA, released the research based on gene mapping. It’s an innovative method which gives better and more accurate results as it consists in creating gene maps which present the position of relative genes on a chromosome.
RESEARCH SUBJECT AND METHOD
The research included a description of a five-generation North American family of European origin. The samples obtained from 13 family members were inspected through gene mapping by using the technology Affymetrix GeneChip Mapping 10K and analyzed in terms of the presence of mutations responsible for limb development in an early embryonic stage. Those mutations are called PITX1 and TBX4. Both mutations PITX1 and TBX4 were identified on chromosome 5q31 among all tested patients, where PITX1 is a homeobox gene.
HOMEOBOX GENES

THE RESULTS
The subject family was affected mainly by predominant idiopathic clubfoot, where the defect was identified as an autosomal-dominant condition with incomplete penetrance. The youngest child was affected by clubfoot on both feet, but was also diagnosed with bilateral foot preaxial polydactyly, right-sided tibial hemimelia and had an additional skin tag near the ear canal entrance. It was at the same time the most difficult examined case in the family. Five family members were affected by clubfoot, three of whom had increased severity on the right, one boy had left clubfoot. Two subjects had bilateral flatfoot (pes planus) – the talus bones were positioned obliquely. One person had developmental hip dysplasia, and two of family members had patella hypoplasia (which affects knee joint functioning). Other family members had other lower limb defects. The research revealed a curiosity – healthy women were mutation carriers, which could explain the fact that less females are born with clubfoot. Men couldn’t pass on this defect, even if themselves they were affected by it.
MICRODUPLICATIONS
It was proved that mutations in PITX1 and TBX4 genes can lead to reduction in lower limb musculature and classic clubfoot phenotypes in children and mice affected by clubfoot, therefore it cannot be excluded that mutation of these exact genes is the reason for isolated clubfoot cases (patients with no other genetic defects). Unfortunately, the same mutations can be associated with syndromic long bone growth defects. The research on TBX4 gene showed the presence of microduplications which impact on the development of the defect. There is no doubt that the research on the genes discovered by the Americans should be continued and clarified. We’re beginning to see the light at the end of the tunel if it comes to the discovery of the cause of clubfoot.
ABERDEEN – LIMK1
The research conducted by dr Martin Collinson and the scientists at the University of Aberdeen showed that one of the genes, called LIMK1, undergoes an overexpression during embryogenesis and is present in all neurons in a developing neural tube at E11.5, that is the 11.5 mouse embryonic day. A part of the research was conducted on mice as they have a phenotype similar to humans, which gives reliable results. The gene LIMK1 encodes one of kinases (serine-threonine kinase) called LIM-domain kinase 1 ( Limk1). This kinase phosphorylates (introduces) the phosphoric acid radicals to certain chemical compounds, in this case to cofilin (a protein), slowing down its actin filament (protein fibers occuring in cytoplasm) depolymerisation activity. In that way, overexpression of LIMK1 would be predicted to inhibit actin turnover – actin is a protein found essentially in cytoskeleton (which takes part, among other things, in keeping and changing the cell’s shape and its movements). According to this phenomena, the scientists proved that motor neurons of the PMA mouse (PMA – peroneal muscular atrophy) with LIMK1 overexpression have high levels of p-cofilin and extend axons poorly compared with a wild mouse. In vivo, the most serious consequence of this appears to be the loss of the peroneal nerve due to its severe atrophy.
SUBJECT, METHOD AND RESULTS OF THE RESEARCH
Process, method and results of the research
To determine empirically if LIMK1 overactivation can result in neuronal defect, the gene activity was mimicked “in ovo” (that is in chicken egg embryos) pharmacologically by using jasplakinolide in order to slow down actin turnover. Jasplakinolide-treated limbs presented a delayed or repressed develoment of the dorsal-projecting axons, which shows that LIMK1 pathway dysregulation may interrupt peroneal nerve growth. These results were confirmed afterwards by additional electroporation in neurons of sciatic nerve lateral motor column (LMC) in ovo in order to induce LIMK1 overexpression during “in vivo” development of the chicken embryo.
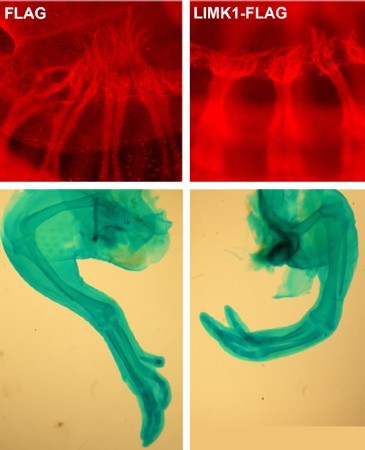
The spontaneous autosomal recessive mutant of mouse’s ‘peroneal muscular atrophy’ (PMA) is a reliable morphological model of human clubfoot. A PMA mouse doesn’t possess dorsal (peroneal) branches of the sciatic nerves. In this research, the original developmental defect was identified as a reduced growth of sciatic nerve lateral motor column (LMC) neurons leading to the conclusion that a static nerve declines or reduces its size along with the whole associated nerve network, causing peroneal muscular atrophy. Genetic and molecular analyses showed that the mutation acts in the EphA4–Limk1–Cfl1/cofilin–actin pathway. In the chicken, both experimental increase of Limk1 kinase using electroporation and pharmacological inhibition of actin turnover led to defects in spinal neuron development and gene path-finding, and mimiced the clubfoot defect.
IS GENETICS IMPORTANT?
Why is genetic research so important? The answer seems to be obvious – if we know the cause, it will be much easier to predict this defect and use more efficient treatment. Despite the fact that new genes, mutations and combinasions are discovered, we still haven’t received a clear-cut answer. Still, it is essential to keep trying to solve this genetic puzzle.

Clubfoot is a complex disorder meaning that more than one gene as well as environmental factors will be discovered to play a role in its etiology. Identifying the genes for clubfoot will allow for improved genetic counseling and may potentially lead to new and improved treatment and preventive strategies for this disorder.

This is, hopefully, another piece in the puzzle of what causes club foot in humans.
RACIAL FACTORS
It seems that ethnic background is also important if it comes to a genetic basis for clubfoot. According to the research conducted by C.S. Chung in 1948-1958 in Hawaii, where the Hawaii population was examined along with its racial variations (Hawaii, Chinese, Korean, Philippine, Puerto Rican and other oriental combinations) in terms of clubfoot, metatarsus varus and calcaneovalgus foot prevalence, the highest clubfoot prevalence was observed among the Hawaii (Caucasians were taken as the base race) and biracial couples (with one Hawaii parent), while among other oriental combinations the risk is lower. As many as 7 per 1000 children affected by clubfoot were Hawaii, whereas among the Chinese the proportion is only 0.39 per 1000 births. The prevalence of clubfoot in terms of racial and genetic differences is still inconclusive. It’s very difficult to establish the fact mostly due to a little base population, just like in the research of Jianhui Wang in 1988 or because of a low percentage of other nations and their constant migration. At the same time, we can’t assume that race or ethnic background is insignificant if it comes to clubfoot inheritance.
| INCIDENCE/1000 LIVE BIRTHS | COUNTRY | DATA SOURCE |
|---|---|---|
| 3.49 | Australia: Aboriginal | M. Carey, C. Bower i inni (2003): “Talipes equinovarus in Western Australia. Paediatric Perinatal Epidemiology” |
| 1.11 | Australia: Caucasian | M. Carey, C. Bower i inni (2003): “Talipes equinovarus in Western Australia. Paediatric Perinatal Epidemiology” |
| 1.6 | Belgium | R. Paton, A. Fox i inni (2010): “Incidence and aetiology of talipes equino-varus with recent population changes.” |
| 1.2 | Denmark | M. Krogsgaard, P. Jensen i inni (2006): “Increasing incidence of club foot with higher population density: incidence and geographical variation in Denmark over a 16 year period.” |
| 6.8 | Hawaii | F. Dietz (2002): “The genetics of idiopathic clubfoot. Clinical Orthopaedics and Related Research” |
| 0.9 | India | R. Mittal, A. Sekhon i inni: “The presence of congenital orthopaedic anomalies in a rural community.” |
| 0.87 | Japan | H. Yamamoto (1979): “A clinical, genetic and epidemiologic study of congenital club foot.” |
| 2 | Malawi | N. Mkandawire, E. Kaunda (2004): “Incidence and patterns of congenital talipes equinovarus (clubfoot) deformity at Queen Elizabeth Central Hospital” |
| 2.7 | Papua New Guinea | A. Culverwell, C. Tapping (2009): “Congenital Talipes Equinovarus in Papua New Guinea: a difficult yet potentially manageable situation.” |
| 0.76 | Philippines | J. Aguilar (nd): “Ponseti method for treating clubfoot in older children and children with previous unsuccessful subtalar releases: short term results.” |
| 1.4 | Sweden | H. Wallander, L. Hovelius i inni (2006): “Incidence of congenital clubfoot in Sweden.” |
| 1.2 | Uganda | S. Pirani, E. Maddumba i inni (2009): “Towards effective Ponseti clubfoot care: the Uganda Sustainable Clubfoot Care Project.” |
| 1 | USA | F. Dietz (2002): “The genetics of idiopathic clubfoot.” |
OGÓLNE RYZYKO WYSTĄPIENIA STOPY KOŃSKO-SZPOTAWEJ JEST TAKIE SAMO DLA CAŁEJ POPULACJI.
W PRAKTYCE OZNACZA TO, ŻE NAWET GENETYCZNIE ZDROWA PARA MOŻE MIEĆ DZIECKO Z TĄ WADĄ,
ALE RÓWNIEŻ PARA OBCIĄŻONA MOŻE MIEĆ DZIECKO BEZ WADY
(ACZKOLWIEK RYZYKO JEJ WYSTĄPIENIA W TAKIM WYPADKU JEST WYŻSZE.)
STATISTICS
Much research concerning genetic causes of clubfoot suggests that certain results are consistent regardless of their place of performance and environmental factors. You can find a simple summary below.
- 2:1
- The defect occurs TWICE AS OFTEN in boys than in girls
- 50%
- approximately 50% of cases are BILATERAL
- NEXT
- if parents don’t have clubfoot but they have a child affected by it, their future children have a 3% chance of having the same foot defect
- ONE
- if one parent is affected by clubfoot, there’s a 3,57% chance that their child will have the same abnormality
- TWO
- if both parents have clubfoot, they have a 10-15% chance that their child will be affected by it as well
- TWINS
- in the case of monozygotic twins, there’s a 32,5% chance that this deformation will affect both siblings compared to 2,9% among dizygotic twins
- I DEGREE
- there’s an approxilately 3% risk of clubfoot among first degree relatives (relations parent-child or sibling-sibling)
- II DEGREE
- there’s an approxilately 0,5-0,6% risk among second degree relatives (grandparents, aunt, uncle)
- III DEGREE
- there’s an approxilately 0,2% risk among third degree relatives (cousins)
SOURCES
RESEARCHES & STUDIES:
1. Idelberger K.: “Die Zwillingspathologie des angeborenen Klumpfußes.”, Stuttgart, 1939
2. Chung C.S. et al.: “Genetic and Epidemiological Studies of Clubfoot in Hawaii: Ascertainment and Incidence.”
3. Tabel data
4. Additional table data
5. Wang J. et al.: “The Role of Major Gene in Clubfoot.”
6. Palmer R. et al.: “The genetics of talipes equinovarus.”, The Journal of Bone & Joint Surgery (JBJS), 1964
7. Palmer R. et al.: “Studies of the inheritance of idiopathic talipes equinovarus.”, 1974
8. Wynne-Davies R. et al.: “Family studies and the cause of congenital clubfoot.”, The Journal of Bone & Joint Surgery (JBJS), 1964
9. Wynne-Davies R. et al.: “Aetiology and interrelationship of some common skeletal deformities.” 10. Yamamoto H.: “Clinical, genetic and epidemiologic study of congenital club foot.”
11. Kurcek A.: Gene mapping 1
2. Gurnett C.A., Dobbs M.B. et al.: “Asymmetric Lower-Limb Malformations in Individuals with Homeobox PITX1 Gene Mutation.”
13. Alvarado D.M. et al.: “Familial Isolated Clubfoot Is Associated with Recurrent Chromosome 17q23.1q23.2 Microduplications Containing TBX4.”
14. Alvarado D.M. et al.: “Pitx1 haploinsufficiency causes clubfoot in humans and a clubfoot-like phenotype in mice.”
15. Alvarado D.M. et al.: “Copy number analysis of 413 isolated talipes equinovarus patients suggests role for transcriptional regulators of early limb development.”
16. Gurnett C.A., Dobbs M.B. et al.: “Genetics of Clubfoot.”
17. Weymouth K.S., Dobbs M.B. et al.: “Variants in Genes That Encode Muscle Contractile Proteins Influence Risk for Isolated Clubfoot.”
18. Alvarado D.M. et al.: “Deletions of 5′ HOXC genes are associated with lower extremity malformations, including clubfoot and vertical talus.”
19. Collinson J.M. et al.: “The developmental and genetic basis of ‘clubfoot’ in the peroneal muscular atrophy mutant mouse.”
20. Protein kinase
21. Phosphorylation
22. Ostrowska Z., Moraczewska J.: “Cofilin – a protein controlling dynamics of actin filaments” 23. Filamenty
24. Myofilaments
25. Actin
26. Holzinger A.: “Jasplakinolide: an actin-specific reagent that promotes actin plymerization.”
27. Yong B.C. et al.: “A systematic review of association studies of common variants associated with idiopathic congenital talipes equinovarus (ICTEV) in humans in the past 30 years.”
28. Basit S. et al.: “Genetics of clubfoot: recent progress and future perspectives.” 29. First Gene For Clubfoot Identified
PHOTOS & GRAPHICS:
1. Washington University in St. Louis
2. DNA
3. University of Aberdeen
4. Elektroporation image: Collinson J.M. et al.: “The developmental and genetic basis of ‘clubfoot’ in the peroneal muscular atrophy mutant mouse.”
5. Matthew B. Dobbs, MD
6. Martin Collinson


